Neuroimage. 2017 Jan 1;144(Pt A):128-141. doi: 10.1016/j.neuroimage.2016.09.049. Epub 2016 Sep 22.
Moradi E1, Khundrakpam B2, Lewis JD2, Evans AC2, Tohka J3.
Author information
- Department of Signal Processing, Tampere University of Technology, Tampere, Finland.
- McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, Canada.
- Department of Bioengineering and Aerospace Engineering, Universidad Carlos III de Madrid, Avd. de la Universidad, 30, 28911, Leganes, Spain; Instituto de Investigacion Sanitaria Gregorio Marañon, Madrid, Spain. Electronic address: jtohka@ing.uc3m.es.
Abstract
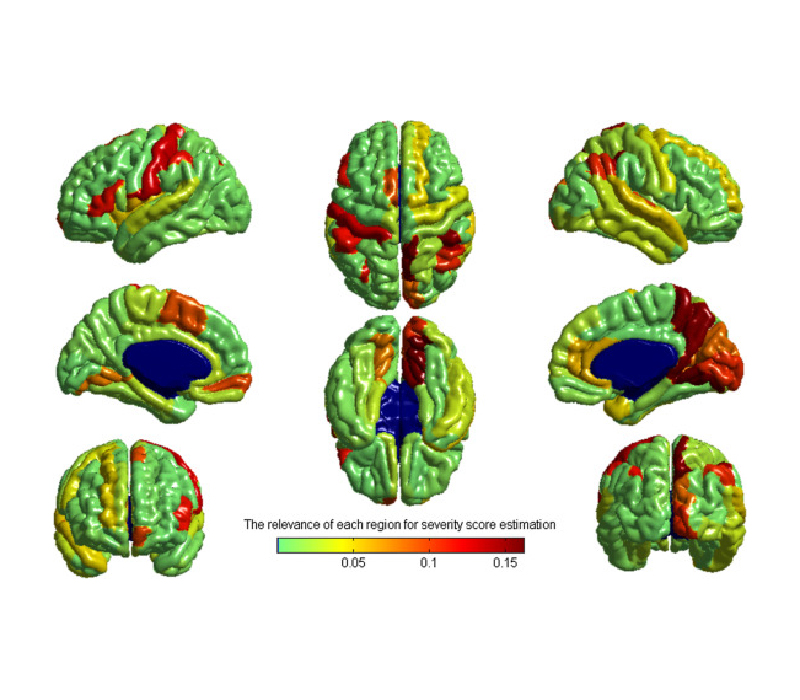
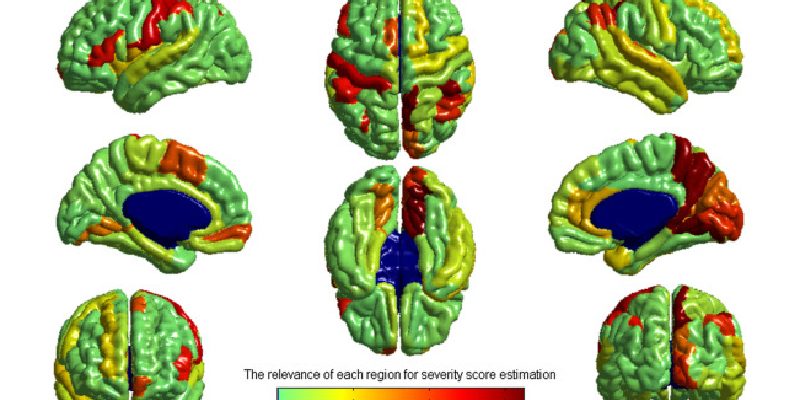
Machine learning approaches have been widely used for the identification of neuropathology from neuroimaging data. However, these approaches require large samples and suffer from the challenges associated with multi-site, multi-protocol data. We propose a novel approach to address these challenges, and demonstrate its usefulness with the Autism Brain Imaging Data Exchange (ABIDE) database. We predict symptom severity based on cortical thickness measurements from 156 individuals with autism spectrum disorder (ASD) from four different sites. The proposed approach consists of two main stages: a domain adaptation stage using partial least squares regression to maximize the consistency of imaging data across sites; and a learning stage combining support vector regression for regional prediction of severity with elastic-net penalized linear regression for integrating regional predictions into a whole-brain severity prediction. The proposed method performed markedly better than simpler alternatives, better with multi-site than single-site data, and resulted in a considerably higher cross-validated correlation score than has previously been reported in the literature for multi-site data. This demonstration of the utility of the proposed approach for detecting structural brain abnormalities in ASD from the multi-site, multi-protocol ABIDE dataset indicates the potential of designing machine learning methods to meet the challenges of agglomerative data.
Copyright © 2016 Elsevier Inc. All rights reserved.
PMID:27664827 | DOI:10.1016/j.neuroimage.2016.09.049