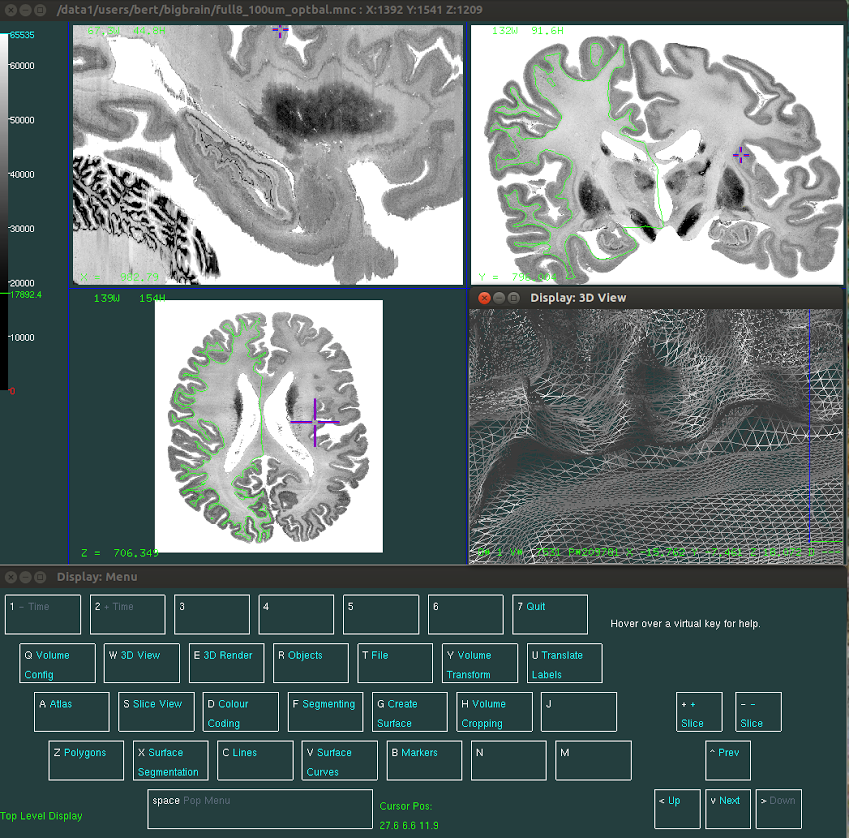
Display is designed to visualize and manipulate three dimensional objects, such as human cortical surfaces and sulcal curves. It also supports the visualization and segmentation of 3D and 4D volumetric images such as MRI, PET, and CT. It was originally written by David MacDonald as part of his thesis research while a student at the McConnell Brain Imaging Centre. MCIN staff are continually improving Display, with new features and bug fixes being added regularly.
Visualization Features Display supports powerful visualization features for both surface-based and volumetric data. Segmentation Features Display allows a researcher to annotate structural features on either a surface or a volumetric dataset. Documentation & Source Code Display is an open source project. Documentation and source code are available at https://github.com/BIC-MNI/Display. We welcome feature requests and bug reports through Github. The Display manual is actively updated with new features being added to Display.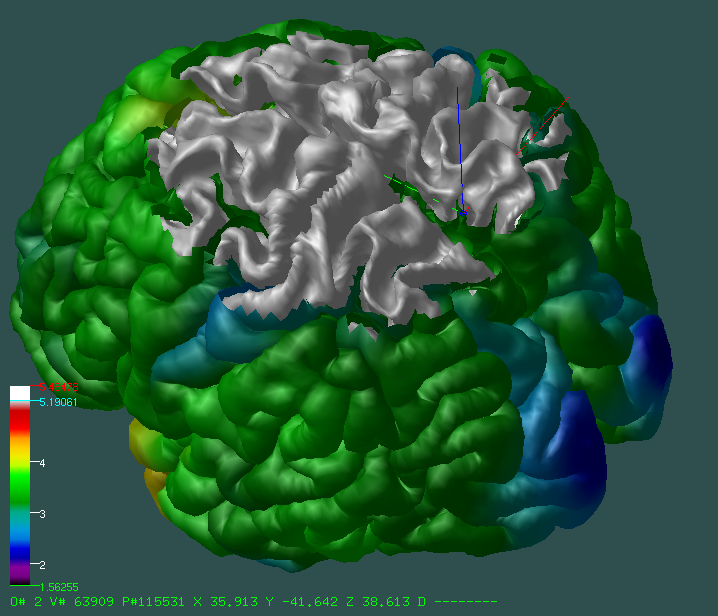
Visualization Features
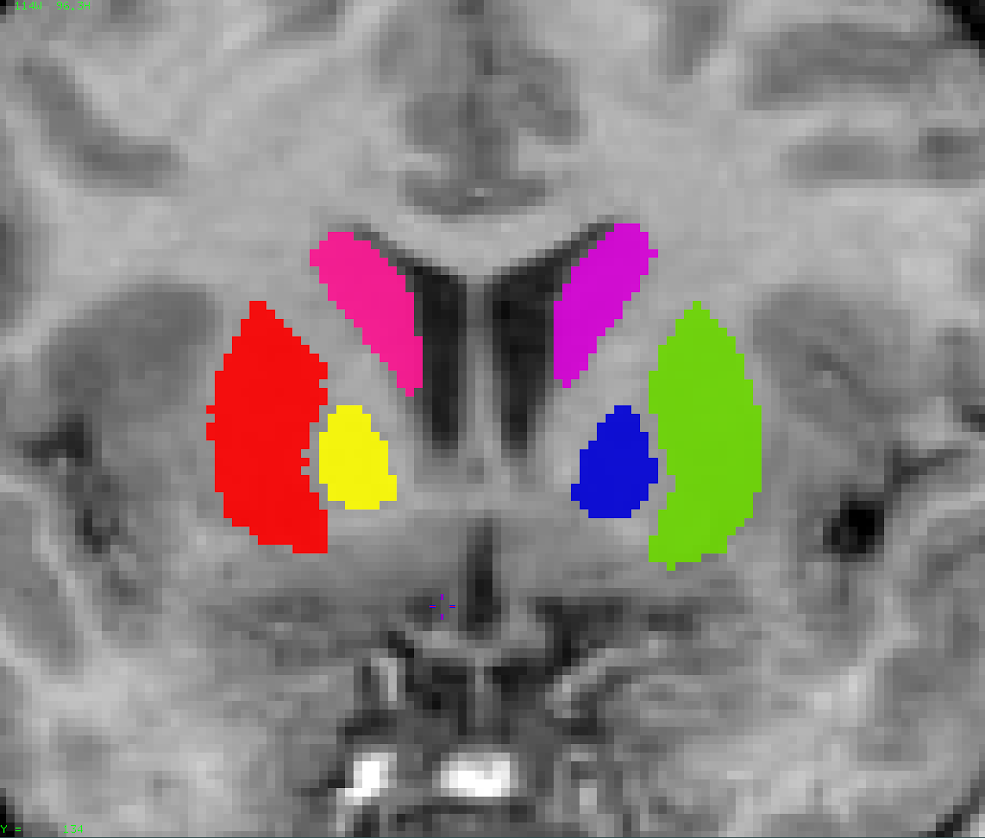
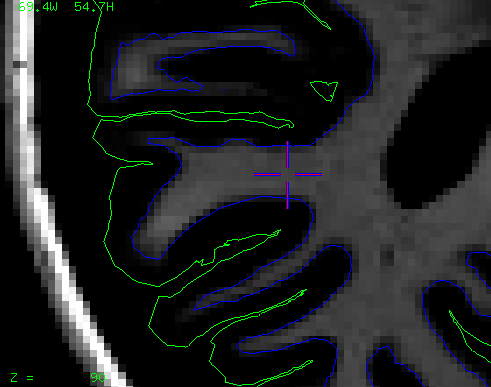
Segmentation Features
Documentation and Source Code